ResourceLife Sciences
Fast Turnaround Times and Improved Data Quality for Microbial Whole-Genome Sequencing with a Novel, Single-Tube Enzymatic Fragmentation
Fast Turnaround Times and Improved Data Quality for Microbial Whole-Genome Sequencing with a Novel, Single-Tube Enzymatic Fragmentation
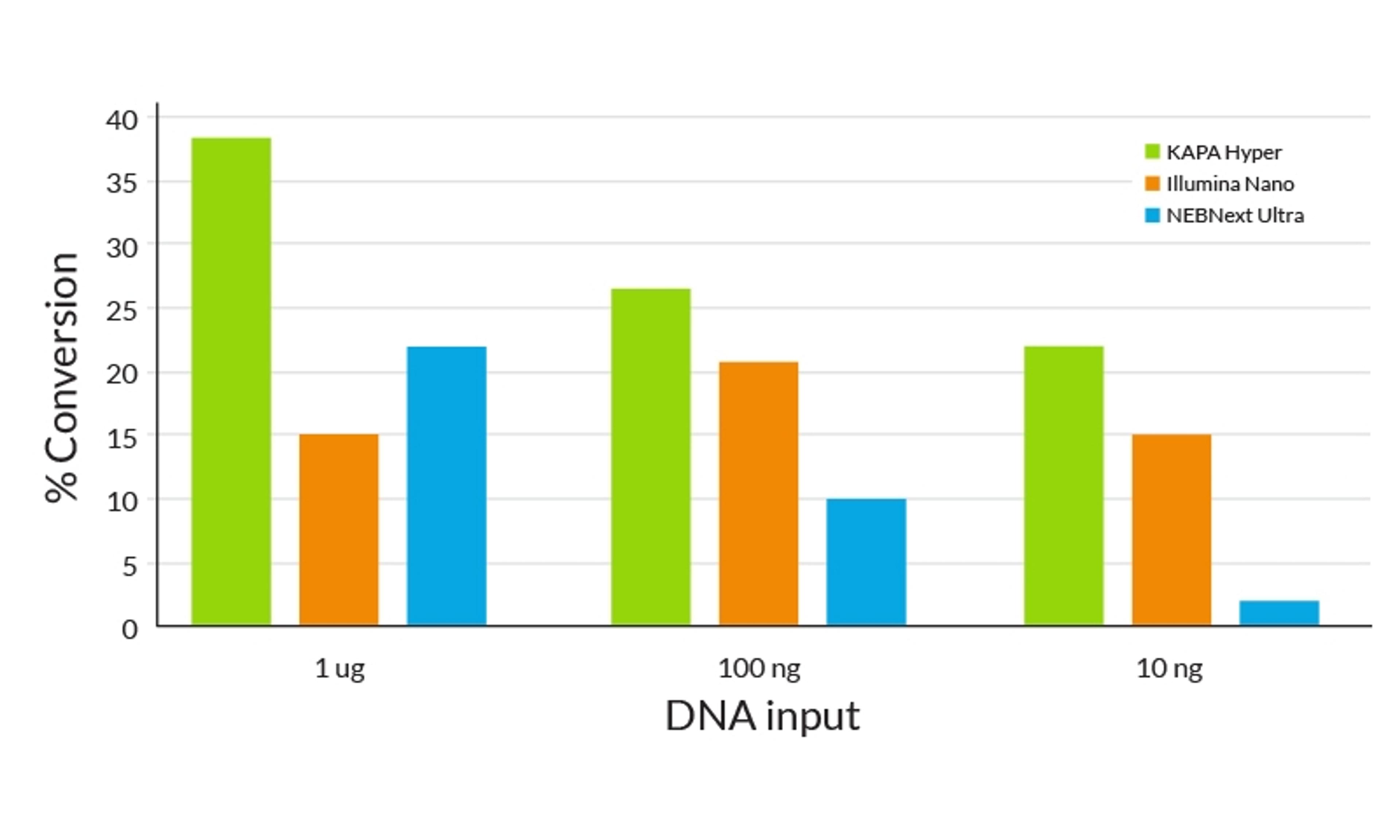
29 Dec 2015Next-generation whole genome sequencing of microbes demands rapid, robust, and scalable library construction workflows, capable of generating high-quality sequence data across a wide range of genome sizes, complexities and genomic GC content. This application note describes a streamlined library preparation method that results in minimal bias, high uniform coverage, and facilitates de novo assembly of microbial genomes.