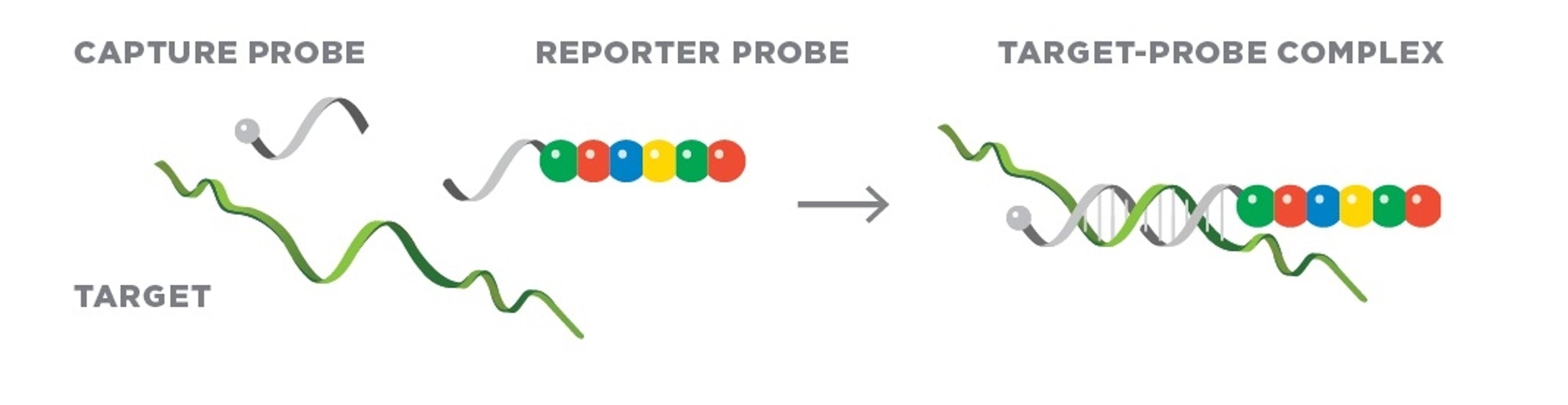
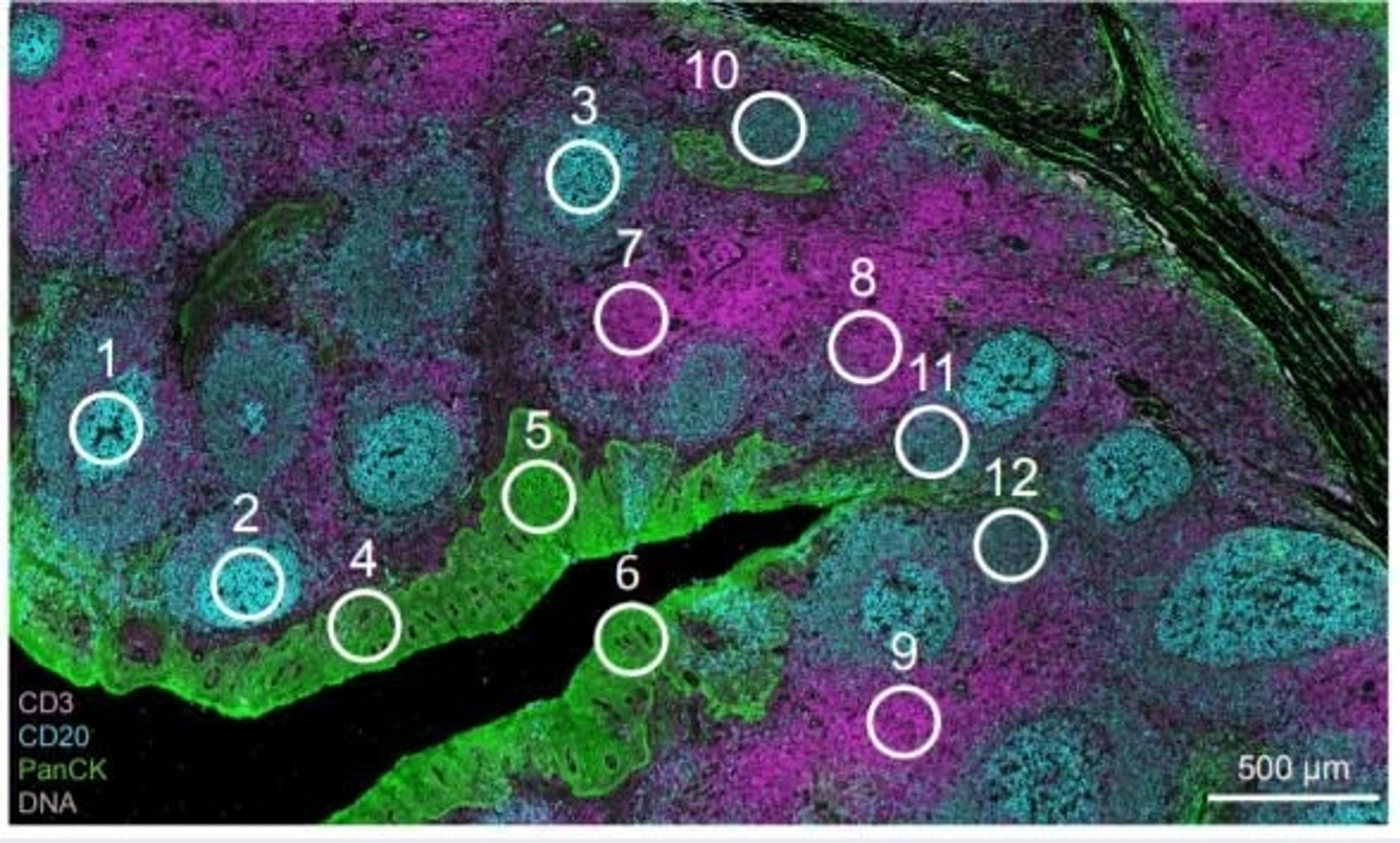
How spatial biology is unveiling new dimensions in research
Dr. Ana de Oliveira from the Spatial Biology Core at UVA discusses the potential of the field to supercharge research across sectors
23 Jul 2024

Dr. Ana de Oliveira, Director of the Spatial Biology Core and Assistant Professor of Pathology at the University of Virginia.
Spatial biology describes the analysis of biological systems in terms of its key components (such as gene and protein expression) in multiple dimensions. Biology happens in three dimensions. Consider embryonic development, in which multiple rounds of gradient signaling activate genes in specific locations to divide and subdivide the body into its final form1. Or consider cancer: oncogenic progression is a spatial process that involves tissue reorganization and the potential for movement to new areas of the body2. Understanding these processes could help us to identify key targets for new anti-cancer drugs, and this principle could also be applied to many other diseases.
With the above examples in mind, it may come as no surprise that the approach affords previously untold opportunities for researchers. “It is the first time we have been able to contextualize analyte expression with its location within tissues on a large scale,” says Dr. Ana de Oliveira, Director of the Spatial Biology Core (SBC) and Assistant Professor of Pathology at the University of Virginia (UVA). “This is unprecedented in the history of science.”
Transitioning from single cells to spatial insights
De Oliveira describes spatial biology as a progression from single-cell analysis approaches. “Single-cell analyses have revolutionized research, but they can’t provide information on cellular localization and the impact of rare cells in tissue microenvironments. Spatial biology can fill these gaps,” she explains. Access to such information could help to supercharge research in areas from drug discovery to disease diagnosis and treatment.: “In pathological research, spatial biology can help us to identify cells that impact disease processes and differentiate cells that respond and do not respond to treatments” de Oliveira shares. “This knowledge could help us to improve outcomes for patients.”
These approaches also tend to extract large amounts of information from small samples, including organoids, cell pellets, and blood smears. The result: potential analysis of rare samples that could not be subjected to study by other methods.
The Spatial Biology Core’s integral role
The SBC was launched in July 2022 to provide researchers at UVA and beyond with access to cutting-edge technology for spatial biology applications, including large-scale spatial genomics and spatial proteomics. The support offered by the SBC covers all key areas of the spatial biology pipeline, including single-cell analyses, spatial RNA sequencing, high-resolution microscopy, and quantitative image analysis. The center also offers scientific consultations and lab space rental for teams that require these services.
“We have a complete infrastructure that supports all stages of spatial analyses across multiple facilities,” de Oliveira says. “The first stage is sample preparation (including block preparation, production of tissue sections, and sample biorepository), which is conducted at the Biorepository and Tissue Research Facility and Research Histology Core. The Advanced Microscopy Facility and Genome Analysis and Technology Core then support quality control steps, such as the morphological assessment of tissues and RNA integrity and sequencing. Finally, downstream data analysis is supported by collaborating with the Bioinformatics Core.”
These offerings are enabled by an impressive range of spatial instruments. “Our instruments include the GeoMx® Digital Spatial Profiler and CosMx™ Spatial Molecular Imager from NanoString and Visium CytAssist from 10x Genomics to run standard and high-definition slides,” de Oliveira explains. “We also have the NanoString nCounter® to conduct multiplex analyses in bulk, the IsoPlexis from Bruker for single-cell proteomics, the Olympus BX53 microscope for infrared analyses, and the ZellScanner One from Canopy-Bruker for chip cytometry.”
However, even with this arsenal to hand, spatial biology studies are not without their challenges. In de Oliveira’s words, “Every new project is a challenge. The first challenge is establishing the experimental design. Experiments must be designed to avoid bias in detecting the analyte(s) of interest. The other key challenge is data analysis. These analyses generate vast amounts of data (about 200 gigabytes of data per 80 fields of view using CosMx™) and the associated computational work can be very challenging. We hope to make an affordable analysis package available to users in the next few months.”
Spatial biology in real-word applications
The SBC recently collaborated with the Instituto Clodomiro Picado in Costa Rica and further institutes within the University of Virginia to publish the first study applying spatial biology approaches to the field of toxicology3.
“We used spatial proteomics to understand the inflammatory response induced during tissue damage induced by snake venom from the Russell’s Viper,” de Oliveira explains. The team monitored 48 proteins involved in apoptosis, inflammation, and tissue regeneration, and identified changes in macrophage phenotypes and their infiltration into venom-treated tissue over time. Apoptosis and extracellular matrix markers were also increased in regions treated with venom, particularly after 24 hours post-injection.
“It was fascinating to monitor all of these processes in unison in this unique experiment,” de Oliveira says. “Studies like this are essential to map biological events and generate insights into how to treat local injuries and systemic disorders. An improved understanding of the physiological processes involved in tissue regeneration could also drive the development of new approaches to treat diseases that affect regeneration of the skin.”
Beyond venom, de Oliveira is also leading a project to study tissue density in breast cancer development with the American Cancer Society and the University of Virginia’s Cancer Center. “I am also collaborating on other studies,” she explains. “But I can’t give more details about those just yet!”
How spatial biology is pioneering progress
Spatial biology is an evolving field that promises previously untold potential for researchers in a myriad of fields. To realize this potential, researchers will have to use their expertise to overcome current hurdles, such as the volume of data produced and potential for errors during manual stages of the process. Answers to these problems might lie in artificial intelligence-based analysis and automation (particularly of stages like sample preparation), respectively.
For the SBC, further innovation may come with the acquisition of new instruments. “We are currently waiting for the CosMx™ Human 6K Discovery Panel, which will drastically increase our coverage of different molecular targets and signaling pathways,” de Oliveira explains. “We also look forward to improvements in data analysis platforms, customized probes for sets of mutations, isoforms, and post-translational modifications, and equipment that allows for the integrated analysis of proteins and RNAs on the same slide.”
The overall hope is to reach a point where spatial biology is used in routine clinical applications to support diagnosis and the provision of personalized treatment. But de Oliveira wants to take things even further. “Our mission is to democratize science and new technologies and make them accessible to all,” she says. “We want to improve quality of life for researchers and patients alike.”
References
Slack J., Establishment of spatial pattern. Wiley Interdiscip Rev Dev Biol (2014).
Seferbekova Z., et al., Spatial biology of cancer evolution. Nat Rev Genet (2022).
de Oliveira A.K., et al., Mapping the Immune Cell Microenvironment with Spatial Profiling in Muscle Tissue Injected with the Venom of Daboia russelii. Toxins (Basel) (2023).