Bringing spatial biology to the clinic: “A new lens on cancer biology”
Spatial transcriptomics pioneer, Dr. Arutha Kulasinghe, shares his latest work in understanding the underlying tumor biology using an integrative multi-omics approach, and how it could inform therapy decisions
11 Oct 2022

Until the recent decade, discussions around the tumor microenvironment (TME) between researchers and clinicians alike were largely limited to understanding hallmark features such as the presence of immune cells, stromal cells, blood vessels, and the presence of an extracellular matrix, but with little insight into whether this intricate and continuously evolving biology plays an active role in cancer progression. Fast forward to present day, and the development of high-throughput spatial omics techniques and technologies has brought with it significant insights into the complex interplay between the tumor and its immediate environment. It is now believed that a dynamic and reciprocal relationship exists between the two, supporting cancer cell survival, local invasion, and metastatic dissemination.1,2
Dr. Arutha Kulasinghe is a Senior Research Fellow and principal investigator at the University of Queensland, whose work with spatial transcriptomics has contributed to leading studies in head and neck, lung, and skin cancer.3-7 As the lead of the Clinical-oMx Lab, his research aims to further understand the pathobiology of patient tumors using an integrative multi-omics approach, whereby multiple datasets from different ‘omics’ such as transcriptomics, proteomics, and metabolomics are combined to interrogate tissues and identify potential diagnostic, prognostic, and therapeutic biomarkers.
In this SelectScience® article, we speak with Dr. Kulasinghe to further explore the insights gleaned from his work on spatial biology, how multi-omics is being adopted by the wider medical and scientific community, and what it takes to move from biomarker discovery to the clinic.
From discovery to translational research
Initially, Dr. Kulasinghe and his team began their multi-omics-based research by taking a wider approach to tumor profiling. Their aim was primarily to understand the tumor-immune-cell interactions that underlie a patient’s response to immunotherapies, such as PD-L1 therapy, with the hope of discovering more predictive biomarkers along the way. “Once you have undertaken these bulk assessments of the tissue. What you really want to be able to do is to drill down into the tumor microenvironment and understand the tumor immune contexture,” explains Dr. Kulasinghe. “We've asked the question, on these cohorts of patient samples, can we do a deep dive using spatial biology to understand the underlying transcriptional profiles, the protein profiles, and then derive cohort-level predictive or putative biomarkers associated with therapy response?”
Over time and as the projects evolve, Dr. Kulasinghe has found that patterns can be observed in these data sets across a number of tumor types. Moving from the initial high-plex discovery studies, the next step was to move into the translational space. “We can narrow these large discovery studies down to more targeted panels that we can now run in a high-throughput manner, potentially in the translational and clinical space.” He goes on to explain this same approach can also be applied to archival retrospective cohorts, which can subsequently be validated in clinical trials to further examine the robustness of the tissue signature.
Applications spanning cancer, infectious diseases, and immunology

Put simply, Dr. Kulasinghe describes the deeper insights that multi-omics provides to not only identify the patients that respond to immunotherapy, but also a way of understanding the patients that are resistant. With this information at hand, a clinician could decide whether better drug options are available or explore other ways of priming the immune system.
Speaking of fellow collaborators working in spatial biology, he explains how the wider community is also utilizing the same technologies. “We've never been able to see these tumors or tissues at this resolution before, even normal tissues are exciting to look at. I know collaborators of ours are running projects where they’re using similar methods to observe how normal tissues age, as we can now see the tissue architecture change over the course of many years by examining past archives.”
Similarly, a lot of work has been undertaken with COVID-19, looking at damage to specific tissues. “Examining patients that died of COVID-19, we collected the lungs, brain, and cardiac tissue.8,9 We were able to apply spatial biology approaches to these tissues to really understand questions such as: Where is the virus localizing to in the tissue? What are the cell types that are being infected? What are the neighboring cell compositions? And what are the cells that are being recruited to the site of infection?” says Dr. Kulasinghe. “For the first time, the infectious disease community is excited by spatial biology because they can apply it to other pathogens of interest.”
Using proteomics to validate genomic and transcriptomic data
Of all the ‘omic’ methods used in spatial biology approaches, proteomics serves as perhaps the most vital because it is the closest assay to the clinic – pathologists are already looking at a wide range of standard single colors such as Ki-67, a marker for proliferation. Dr. Kulasinghe explains how pairing it with transcriptomics alone can bring significant insights, “I think being able to do discovery, looking at RNA upfront through transcriptomics, then validating those findings using proteomics is key to being able to really understand what's happening at the cellular level. A lot of proteomics technologies now also support multiplexing, and once you’re running 10-, 30-, or even 50-plex, spatial proteomics becomes a discovery tool almost in itself.”
Dr. Kulasinghe continues to describe how researchers and clinicians are rapidly gathering in this spatial proteomics field to outline it as a way of digitizing a pathologist's assessment of tissue. “It is really exciting for us all. I think if we can develop panels, whether they're metabolic panels, T-cell infiltration, T-cell exhaustion, myeloid panels, or tumor profiling panels, and run them as standard assays on all samples, we can glean a lot of insights, which can then be used to inform on therapy response.”
The ideal spatial biology platform
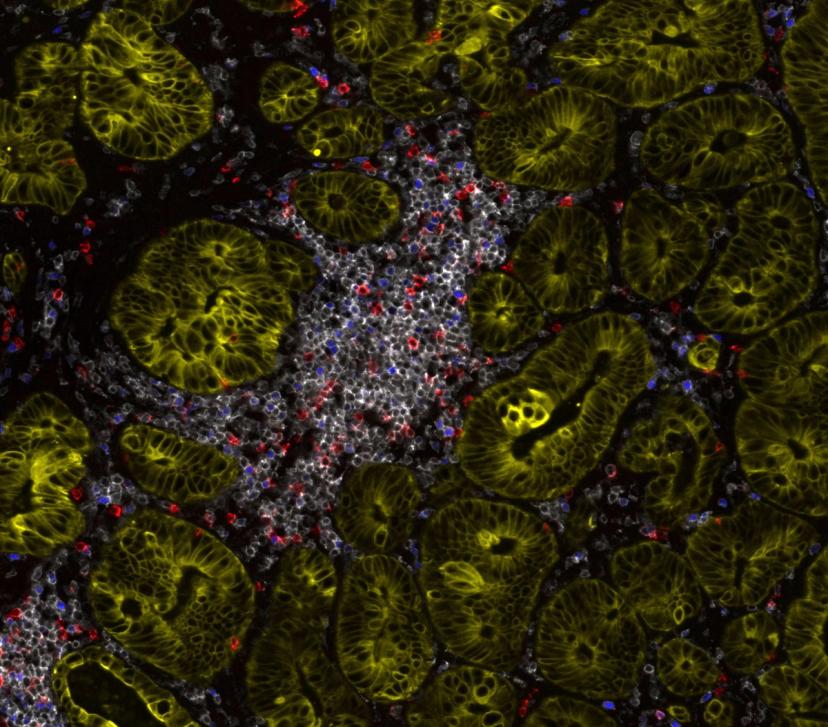
When asked ‘What makes an ideal spatial proteomics platform for translational research?’, Dr. Kulasinghe explains that high-plex, around the 15 to 20 color mark, is ideal when working in the discovery space. With resolution also playing a major factor. “We really want to be able to resolve the signal down to single-cell resolution. If you're able to identify your markers confidently at single-cell resolution, that becomes very powerful because now you can quantify it, compare it, and identify the localization of those markers, the cell types, and the neighborhoods that those markers are present in.”
Alongside resolution and high-plex, flexible panel design and the ability to custom conjugate markers of interest plays a major role in moving to a translational setting. “For clinical projects, we want to be able to use off-the-shelf predeveloped panels that are wet-lab-validated and can be run routinely. Equally, if we had markers of interest developed from discovery studies, and we want to be able to plug those into those panels, then we should have the ability to custom conjugate,” Dr. Kulasinghe states. “With the large and complex data sets generated, having an analytical framework and integrated image analysis software so you can really understand the outputs, identify cell types, count, enumerate, and pinpoint neighborhoods to perform segmentation is important.”
Creating tests of clinical utility
Moving from translational research to the clinic is now ultimately the focus for Dr. Kulasinghe and his team. The challenge lies with taking the broad RNA and protein panels with hundreds of markers and paring these down to smaller, affordable, more targeted panels and validating them for clinical use. “To achieve this, we not only need flexible and innovative platforms, we need to take the pathologists with us. They are key to the translation of this work,” he explains. “We can do this by working with them throughout, rather than taking the final product to them. We need their input to annotate the tissues, to identify the cell types on H&E-stained samples, before we superimpose our multiplex spatial images. When we do that, there's a synergism, which will enable the move to the clinic much faster. Pathologists are going to be critical for this next wave of adoption.”
Obtain whole-slide, high plex biomarker quantitation with the all-in-one spatial biology benchtop platform by RareCyte, Inc.
Meet Dr. Kulasinghe and the Orion platform at the 2022 Multi-Omics meeting in Brisbane, Queensland, December 11–13.
References
Anderson N, Simon M, The tumor microenvironment. Current Biology. 2020. PubMed
Wu Y, Cheng Y, Wang X, et al. Spatial omics: Navigating to the golden era of cancer research. Clinical Translational Medicine. 2022. PubMed
Sadeghirad H, Monkman J, Mehdi A, et al. Dissecting Tissue Compartment-Specific Protein Signatures in Primary and Metastatic Oropharyngeal Squamous Cell Carcinomas. Frontiers in Immunology. 2022. PubMed
Kulasinghe A, Monkman J, Shah E, et al. Spatial Profiling Identifies Prognostic Features of Response to Adjuvant Therapy in Triple Negative Breast Cancer (TNBC). Fronteirs in Oncology. 2022. PubMed
Rad HS, Shiravand Y, Radfar P, et al. Understanding the tumor microenvironment in head and neck squamous cell carcinoma. Clinical and Translational Medicine, 2022. PubMed
Kulasinghe A, Tan CW, Ribeiro Dos Santos Miggiolaro AF, et al. Profiling of lung SARS-CoV-2 and influenza virus infection dissects virus-specific host responses and gene signatures. The European Respiratory Journal. 2022. PubMed
Monkman J, Kim H, Mayer A, et al. IL-2 stromal signatures dissect immunotherapy response groups in non-small cell lung cancer (NSCLC). medXriv. 2021. medXriv
Kulasinghe A, Liu N, Tan CW, et al. Transcriptomic profiling of cardiac tissues from SARS-CoV-2 patients identifies DNA damage. immunology. 2022. Wiley

