Mass photometry: Shining a new light on protein self-assembly
Learn how a novel method for label-free single-molecule characterization is changing our understanding of key biological processes
9 Dec 2022

In this SelectScience® interview, we speak with Dr. Nikolas Hundt, a researcher at the Ludwig Maximilian University of Munich, Germany, to find out how his team is applying mass photometry – a novel label-free technique for quantifying the mass of individual molecules in solution – to shed new light on key biological processes. Hundt shares how his latest research using this approach has generated new insights into the underlying mechanism of protein self-assembly that have challenged and revised existing models. He also highlights the main strengths and limitations of this burgeoning technology and reveals what he hopes for its future.
Measuring molecules with light
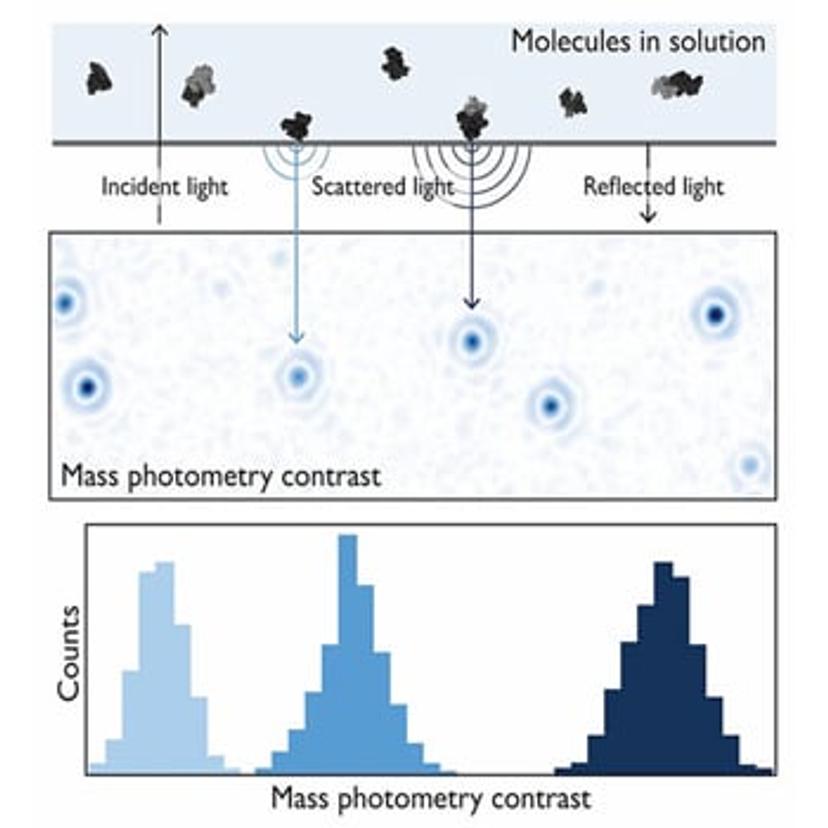
Mass photometry is a single-molecule technique that measures molecular mass by quantifying light scattering from individual biomolecules in solution. During a mass photometry experiment, a sample of proteins or other biomolecules is illuminated with a beam of light. The interference between the light reflected by the measurement surface and the light scattered by a molecule in contact with the measurement surface generates a mass photometry contrast (Fig. 1). The magnitude of this interference scales linearly with molecular mass, and via calibration, the technique enables the mass measurement of biomolecules with high accuracy.
Using this novel technique, researchers can investigate molecular structure, characterize sample heterogeneity, and study biomolecular interactions with high resolution – all without the use of labels. It has already been used in more than 150 published papers to study a diverse range of biomolecules, including proteins, nucleic acids, large macromolecular complexes, nanostructures, and even small viruses such as adeno-associated viruses (AAVs).
Hundt was involved early on with the development of mass photometry during his time at the Kukura group at Oxford University. In 2018, the spin-out Refeyn was founded, a company that aims to share this technology with the global scientific community through its flagship mass photometer, TwoMP. Now, based at the Ludwig Maximilian University of Munich, Hundt continues to push the boundaries of the method’s capabilities, with a focus on protein self-assembly.
A new view of protein self-assembly
In his latest study1, Hundt applied mass photometry to investigate the early assembly steps of actin, a globular protein that transitions between monomeric and filamentous states to mediate many critical cellular processes, including cell motility, cell division, vesicle and organelle movement, and the maintenance of cell shape. Actin has been studied since the 1940s and has played a seminal role in our understanding of biopolymer formation. However, efforts to determine the molecular mechanism underlying actin filament formation have generated several different, at times contradictory, models. Hundt’s study was the first to observe this process at single-molecule resolution. “When I could first see actin nucleation with mass photometry, I was very excited, but I saw a lot more nucleating species than was predicted by the common models,” he shares. “We found that the old models didn't use all the knowledge that was already known about actin – for example, its ATPase activity, the fact that different ends polymerize with different speeds, and so on,” he continues. “So, we tried to interpret all this information in a different way, and then came up with a new model that could fit both the bulk data and the mass photometry data as well.”
The benefits of a mass photometry approach
Until now, the go-to method for understanding actin polymerization has been to measure the bulk signal of a solution via light scattering or fluorescence to determine changes in the number of filaments forming over time. This experimental data is then compared to different kinetic models to find those that best reproduce the data under different conditions and concentrations. “This approach has not only been applied to actin filaments, but to all sorts of protein polymers such as microtubules, α-synuclein, and various amyloids,” says Hundt. However, a major challenge intrinsic to this method is that numerous nucleation models can often fit the same experimental data. “This is where mass photometry came in because for the first time it was possible to look at the species that were present from the very beginning,” he shares. “Since you directly observe and quantify all the individual nucleation species, you can resolve these ambiguities when several models fit the same dataset and understand what is really happening on a molecular level.”
The findings of Hundt’s recent study revised the mechanism of actin nucleation and introduced mass photometry as a quantitative means to study protein self-assembly at the molecular level. “Using mass photometry means you can very easily detect the formation of complexes, and from the size and distribution, you can immediately determine stoichiometries and affinity constants,” he says. “Compared to alternative methods such as size-exclusion chromatography (SEC) or dynamic light scattering (DLS), much less protein is required, and it’s also label-free, so you don't have to use fluorescent dyes or other tags which can disturb the natural behavior of proteins under investigation.”
The experimental simplicity of mass photometry is also a key advantage. “With the way mass photometry instruments are implemented now, the method is very simple and fast,” he continues. “In principle, you can take a sample, put it on a cover slip, and immediately observe your molecules landing.”
Experimental considerations
Despite the many advantages of mass photometry, as with all analytical methods, researchers must keep in mind a couple of limitations when investigating protein self-assembly. Principal among these is the requirement to work in a relatively low concentration regime. “As mass photometry is a single-molecule technique, you have to work with low concentrations, which means that if your proteins have low-affinity interactions, you might not see complex formation,” explains Hundt. “In addition, the size distribution of proteins might not be directly comparable to other techniques such as SEC or DLS. For example, if your protein is a tetramer at high concentrations, it’s likely that it disassembles at one or more of the concentrations used in mass photometry.”
When using complex samples, a lack of specificity can also complicate the analysis of mass photometry experiments. “Although being label-free is a great advantage of the technique, it can also mean that the signal generated is not very specific,” he adds. “You need to have a fairly good idea about what your sample contains because otherwise, it can become hard to interpret your results.”
Future outlooks
The broad applicability of mass photometry, combined with its ability to accurately determine a wide range of species without the need for labels, makes it an extremely appealing method for biomolecular research. “I think it will become a routine method in molecular biology, especially the more automated the whole experiment becomes,” asserts Hundt. “I can also imagine that it will be used more for non-biological samples, such as polymers or macromolecules in the chemical industry.”
Efforts to improve the sensitivity of current mass photometry technology as well as address its limitations are likely to form key areas for future development. “For example, developing surface passivation techniques in order to be able to work at higher concentration ranges, or trying to improve the image analysis in order to see even smaller proteins,” says Hundt. “Finally, I’m excited to see the technique being used in combination with different types of spectroscopy methods, such as Raman spectroscopy. With mass photometry, you can get accurate mass and size information, but if you could also make the signal more specific and identify the chemical content of each signal, it would become even more versatile,” he concludes.
Reference
Hundt N, Cole D, Hantke MF, et al. Direct observation of the molecular mechanism underlying protein polymerization. Science Advances 8(35), eabm7935 (2022). Science Advances

