Design your CRISPR gene editing experiments in as little as five minutes
Discover how the suite of gene editing technologies from Thermo Fisher Scientific is simplifying gRNA and experimental design
1 Nov 2022The toolbox of gene editing solutions available to scientists has evolved over recent years, enabling more efficient editing for a range of applications. While genome engineering is not a recent endeavor, with its beginnings going back decades, science has progressed beyond the use of zinc finger nucleases, towards faster, easier and more efficient techniques such as transcription activator-like (TAL) effector nucleases (TALEN) and CRISPR-Cas9 gene editing methods. One of these techniques, CRISPR gene editing, has emerged as one of humanity’s most powerful technologies with the potential to change the way we create energy, produce food, optimize industrial processing and detect, prevent and cure disease. The next generation of CRISPR-Cas9 tools is now allowing researchers to target almost any gene for modification or deletion/silencing in a highly specific and reliable way.
In this editorial, we look at key considerations for designing a successful and precise CRISPR gene editing experiment. We will also demonstrate how the InvitrogenTM TrueDesignTM Genome Editor software and associated tools from Thermo Fisher Scientific can help support every step in your gene editing workflow.
Things to consider when designing CRISPR experiments
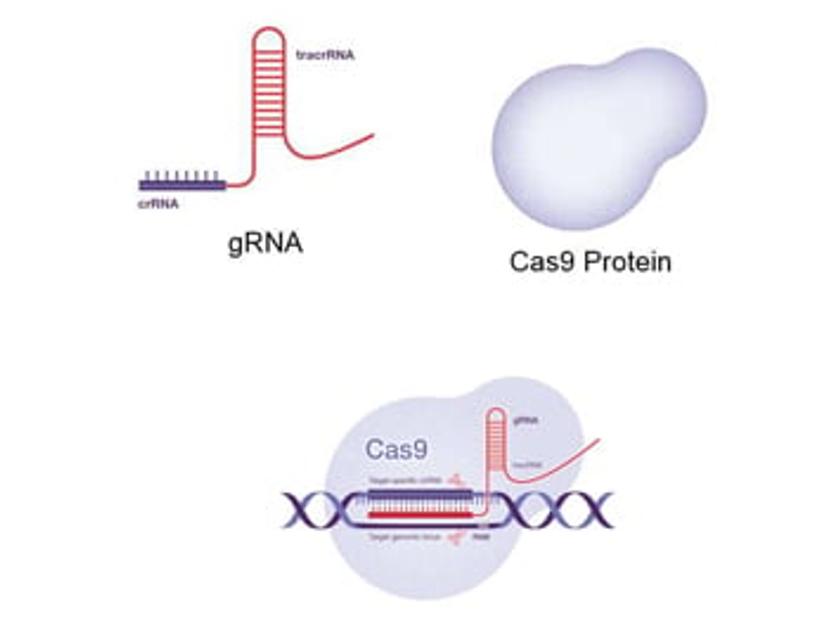
CRISPR gene editing systems contain two key components, a guide RNA (gRNA) and CRISPR-associated endonuclease, with Cas9 commonly adopted. The gRNA contains a user-defined spacer sequence of around 20 nucleotides in length that allows targeting of the Cas protein to a gene of interest. The target DNA sequence is also an important consideration as this must be upstream of a short DNA motif known as the protospacer adjacent motif (PAM) for the gRNA:Cas complex to cut genomic DNA. The selection of an optimal target site, with a consideration of available PAM motifs, as well as the exact matching of gRNA and target sequences, are therefore crucial for gene editing success.
Whether you’re working on a routine CRISPR-Cas9 edit in standard cell lines or trying to achieve a specialist edit in primary cells or induced pluripotent stem cell (iPSC) lines that requires maximum targeting efficiency, each approach requires careful consideration of the experimental design and build. Thermo Fisher Scientific provides a comprehensive suite of gene editing tools to support each stage of the gene editing workflow, from designing guide RNA (gRNA) for your gene target of choice to delivery and downstream characterization.
Before you start your experiment:
- Consider designing and testing three gRNAs for every target.
- Run bioinformatic analysis to choose gRNAs with minimal predicted off-target effects using tools like TrueDesign Genome Editor software.
- For knockout experiments, target early constitutive exons to disrupt the reading frame.
- For knock-in experiments, the cleavage site should be as close to the edit site as possible (<10 bp if possible) and the lengths of the donor homology arms should be optimized. TrueDesign Genome Editor can automatically design custom homology arms for your knock-in experiments.
- Choose a Cas nuclease best suited for the experiment and cell type.
The Invitrogen TrueDesign Genome Editor software takes all of this criterion into account to make designing your gene editing experiment much less stressful.
Download the Invitrogen Genome Editing Resource Guide for additional tips & tricks
The InvitrogenTM TrueGuideTM Synthetic Guide RNAs from Thermo Fisher Scientific offer a predesigned CRISPR sgRNA produced via algorithms that select for high knock-out efficiency with optimal specificity. For custom-made gRNAs, the Invitrogen TrueDesign Genome Editor offers both ease of design and the selection and ordering of the correct reagents.
Designing gene editing experiments using TrueDesign Genome Editor
The TrueDesign Genome Editor software is a free online tool that enables researchers of all experience levels to easily design, select and order reagents for accurate and successful gene editing experiments across five different species including human, mouse, rat, zebrafish and roundworm.
The software supports seven types of gene editing: gene knock-out, gene knock-in, fluorescent tagging, epitope tagging, replacement, deletion, and SNP creation. It follows a simple three-step workflow in which you select your gene and transcript, specify your edit, and then design your CRISPR target from the software’s recommendations. For each design, the software compiles a list of the materials and reagents that will be required for a successful edit, and conveniently makes provision for ordering them from the same source, giving confidence that they will work together.
Gene knock-in : A TrueDesign Genome Editor case study
Whilst gene knock-outs have been achievable for several years, gene knock-ins have remained problematic and elusive because of the low efficiency of homology-directed repair. To be successful, knock-ins depend upon the precise design of donor DNA, the effectiveness of the sgRNA and Cas9 complex, and the efficiency of delivery into the target cells. View this application note on using TrueDesign Genome Editor to improve and simplify the design of gene knock-ins, featuring the introduction of single nucleotide polymorphisms (SNPs) and fluorescent tags into induced pluripotent stem cells (iPSCs) with up to 100% editing efficiency.
As an example of this approach, research published by Kim et al., (2022) in Nature Communications dissects leucine metabolism based on glucose sensing and availability. Using the TrueDesign Genome Editor from Thermo Fisher Scientific to design sgRNA and donor DNA, the team at Yonsei University in Seoul, South Korea, used CRISPR, and TrueCutTM Cas9 Protein v2, to generate LARS1 S1042A knock-in cells and to demonstrate that the enzyme plays an important regulatory role in leucine metabolism in response to glucose starvation. This study demonstrates the power of CRISPR-Cas9 technology to help unravel the complexities of cellular metabolism, given its ability to study, alter, create and re-create highly complex pathways, DNA sequences and genes.
Learn more about the suite of end-to-end gene editing solutions from Thermo Fisher Scientific and more specifically how the Invitrogen TrueDesign Genome Editor could help you when designing your next gene editing experiment.
Reference
Kim, K., Yoo, H.C., Kim, B.G., Kim, S., Sung, Y., Yoon, I., Yu, Y.C., Park, S.J., Kim, J.H., Myung, K., Hwang, K.Y., Kim, S. & Han, J.M. O-GlcNAc modification of leucyl-tRNA synthetase 1 integrates leucine and glucose availability to regulate mTORC1 and the metabolic fate of leucine. Nature Communications 13(1):2904; May 25, 2022. DOI: 10.1038/s41467-022-30696-8. PMID: 35614056; PMCID: PMC9133088.