The potential of immunoproteomics in type 2 diabetes as a diagnostic and prognostic tool to combat bacterial infection
Discover how researchers at Weill Cornell Medical College are using immunoproteomics to identify biomarkers that help better understand and manage diabetic infections
28 Nov 2023

Just under half a billion people are living with diabetes worldwide and the number is projected to increase by 25% in 20301. It has long been established that there is a link between genetics and diet with the development of diabetes, but more recent research has also found a connection to bacteria. To further understand this connection, the proteomics core facility at Weill Cornell Medical College is using immunoproteomics in type 2 diabetes research to identify specific antibodies that correlate with the progression of diabetic infections. This information can then be used to put in place targeted measures against the bacteria involved and may provide insights into disease pathogenesis and healthy prognosis.
Proteomics plays an essential role in the identification and quantification of proteins for the analysis of disease-specific markers. Alongside immunoproteomics, these approaches are used to better understand the molecular mechanism behind various human diseases. In this article, Professor Frank Schmidt, Associate Professor of Biochemistry and Director, Proteomics Core, Weill Cornell Medicine–Qatar, discusses the link between bacterial infection and type 2 diabetes. Prof. Schmidt also explores how his team is using novel protein array technology to accelerate the discovery of autoantibodies produced in response to diabetic infections.
Type 2 diabetes puts patients at greater risk of bacterial infection
It is established that type 2 diabetes affects the immune system. More than 90% of diabetes patients have type 2 diabetes mellitus, a metabolic disease marked by hyperglycemia, which increases the risk of systemic infections2. “If an immune-compromised person with diabetes gets a bloodstream infection with bacteria such as Staphylococcus aureus this can lead to a host of medical complications,” explains Prof. Schmidt. “Due to increased level of glucose caused by hyperglycaemia, bacteria in the blood or on the skin of diabetes patients can excess glucose more efficiently than bacteria in healthy patients. This proliferates the spread of the bacteria and results in a higher chance of severe infection.”
As well as diabetes patients being at greater risk of hospitalization and mortality from infections, there are also hints that chronic infection with certain bacteria and viruses can trigger diabetes. “We have evidence that suggests virus infection can trigger type 1 diabetes and there is also evidence suggesting that COVID-19 can trigger type 2 diabetes, due to damages to insulin-producing cells3,” says Prof. Schmidt.
The role of immunoproteomics in understanding bacterial infection
To better understand the connection between type 2 diabetes and infection, Prof. Schmidt’s team is using proteomics to investigate the over and under-expressed proteins in a human host and the pathogen. “When it comes to immunoproteomics, we want to study the antibodies which are newly synthesized in the host that target proteins secreted by the bacteria,” explains Prof. Schmidt. Whilst this may seem simple in practice, the study of antibody reactions to bacterial infections can be complex. For example, with COVID-19 you get a clear antibody reaction in response to a single spike protein, but Prof. Schmidt explains that studying antibody reactions to bacteria is much more complex, “Staphylococcus aureus, for example, can secrete up to 500 different proteins, many of which can modulate the immune system.”
By studying the antibody response in healthy subjects, we can elucidate which proteins would be suitable to target and produce potentially therapeutic neutralizing antibodies against these.
Professor Frank Schmidt
Weill Cornell Medicine – Qatar
By understanding the antibody response to bacterial proteins, it may be possible to create therapeutics based on neutralizing antibodies to help treat people with diseases. “Healthy subjects can build up a huge repertoire of antibodies against bacterial proteins. We know from studies that not all of the proteins are targeted, and some are hidden due to the presence of lipid bilayers and glycosylation. By studying the antibody response in healthy subjects, we can elucidate which proteins would be suitable to target and produce potentially therapeutic neutralizing antibodies against these,” says Prof. Schmidt.
Long-term exposure to bacteria or viral proteins can result in chronic inflammation in patients. “We have seen in recent studies that if you have a chronic inflammation for some time, the body will start to produce autoantibodies,” explains Prof. Schmidt. “It is known that sufferers of type 2 diabetes tend to have more pathogens on the skin and are more prone to chronic inflammation. We are currently conducting a study to confirm the link between chronic inflammation and the production of autoantibodies which can then be used to inform future research into potential therapeutics.”
Protein-folding technology: A better way to study antibody responses
Previously the Proteomics Core at Weill Cornell Medicine–Qatar used FLEXMAP 3D for the detection of different types of antibodies. “This is a bead-based method, where we study the proteins in a denaturized form, and then incubate them with patient serum. The disadvantage we found when using this method is that the proteins are not folded correctly,” says Prof. Schmidt. “It’s important that we simulate the situation in the body as closely as possible and correct protein conformation is important for antibody binding studies.”
To overcome this, Prof. Schmidt’s team turned to the KREX® protein-folding technology from Sengenics. “In the beginning, we used one of Sengenics' commercially available chips. We then began production of customized chips with Sengenics so that we could be more specific to our research, and so far, we have promising results,” says Prof. Schmidt.
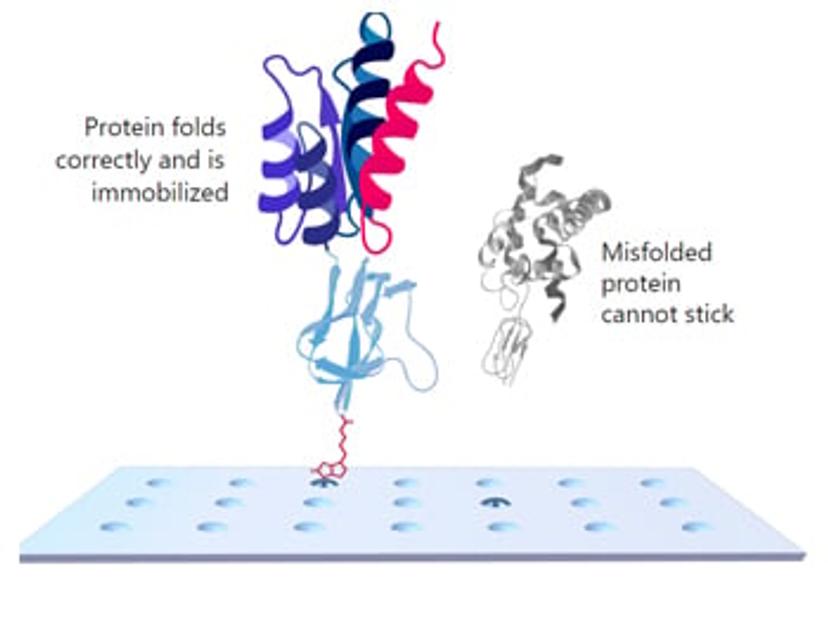
KREX ensures that only correctly folded proteins are immobilized on the array surface. During the assay, when a protein is correctly folded, it co-translationally drives the correct folding of biotin carboxyl carrier protein (BCCP) which then becomes biotinylated. This allows the fusion protein to become attached to the surface of the array. On the other hand, misfolded proteins co-translationally drive misfolding of BCCP. This results in BCCP no longer becoming biotinylated, preventing the misfolded protein from attaching to the array. This ensures each assay is run on correctly folded, full-length, functional proteins which is crucial for specific antibody detection.
Future outlooks
Improvements in proteomics technology and the development of functional protein array technology such as KREX offer vast practical value for advancing our understanding of type 2 diabetes and infection. These developments are also helping accelerate the discovery of new autoantibody biomarkers. Despite these developments, there is still a way to go to bridge the gap between genomics and proteomics. “Proteomics technologies are still behind genomics, and this has something to do with the nature of proteomics. We have roughly 20,000 genes, but we have three to four different million proteoforms. With techniques like mass spectrometry, we can now detect thousands of proteins in the blood, but not yet all the proteoforms,” says Prof. Schmidt.
Prof. Schmidt is optimistic that biomarkers identified using proteomics will continue to have an impact on the discovery of new therapeutics. “We now see new biomarkers coming from the proteomics field. In parallel, we also see more and more autoantibody studies using the KREX technology. There is evidence that autoantibodies can be very useful as a biomarker as they are statistically relevant and robust.”
In the future, as research moves towards precision medicine, Prof. Schmidt hopes that we will be able to combine genomics information with proteomics and immunoproteomics to develop personalized medicines. “Differences between people’s immune response can be the result of differences in individual single nucleotide polymorphisms (SNP) or even hundreds of different SNPs per person. If we can translate these genomic differences into protein or autoantibody information, we can then start to identify possible drug targets which are specific to an individual.”
References
Saeedi, P. et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9th edition. Diabetes Research and Clinical Practice, (2019) Nov: 157:107843. Pubmed
Sohail, M.U. et al. The role of pathogens in diabetes pathogenesis and the potential of immunoproteomics as a diagnostic and prognostic tool. Frontiers in Microbiology, (2022) Nov; 13:1042362. Pubmed
Mallaparty, S. Mounting clues suggest the coronavirus might trigger diabetes. Nature, (2020) Jul; 583:7814. Pubmed

