What next for CRISPR after first therapeutic approvals?
As we celebrate the first regulatory approval of a gene-editing based therapeutic, Casgevy™, for the treatment of sickle cell disease and transfusion-dependent β-thalassemia, we consider the gene-editing challenges that must be overcome before CRISPR gene-editing technology becomes a common treatment modality
12 Feb 2024
Gene-editing techniques possess the unique potential to cure genetic diseases at the molecular level, contrasting with current treatments which tend to rely on alleviating the symptoms of disease. This revolutionary approach offers a one-time, enduring effect instead of a lifelong commitment to managing disease symptoms1. However, challenges of in vivo CRISPR therapeutics currently hinder the potential of CRISPR as a commonplace treatment modality.
With the UK MHRA’s approval of Casgevy (exagamglogene autotemcel - exa-cel) in November 2023, clustered regularly interspaced short palindromic repeat (CRISPR) gene editing has evolved from inception to therapeutic reality in little more than a decade2,3. While this world-first gene therapy treatment represents a significant milestone, we have barely scratched the surface of the potential for gene therapies to revolutionize healthcare.
CRISPR-Cas9 leads the charge
Although the human genome is complex, many common genetic disorders arise from mutations in just a single gene, such as cystic fibrosis, sickle cell disease, Duchenne muscular dystrophy, and Huntington’s disease. There are over 500 monogenic diseases that affect more than 250 million individuals worldwide. DNA sequencing in affected families guides the identification of DNA sequence modifications that could potentially serve as a genetic cure, either by disrupting the function of a harmful or inhibitory gene or by restoring the function of an essential gene4.
This manipulation of the genome is facilitated by gene editing technologies, such as CRISPR and its associated protein (Cas-9). CRIPSPR-Cas9 stands out as the most effective, efficient, and precise method for genome editing when compared to earlier techniques like zinc finger nucleases (ZFN) and transcription activator-like effector nucleases (TALENs)5.
Achieving targeted genomic cleavage relies on two essential components: a homing device and an endonuclease. In the case of CRISPR, a single-stranded RNA (sgRNA) serves as the homing device, guiding the Cas protein (endonuclease) to the intended target site. The recognition process of sgRNA is reliant upon protospacer-adjacent motifs (PAMs), short guanine-enriched sequences at the target site. Cas9's preferred PAM sequence, NGG, is a prevalent motif in the human genome. This facilitates the application of CRISPR technology in medical contexts6.
Casgevy and the code to success
The therapeutic approach of Casgevy involves the use of CRISPR-Cas9 to target the BCL11A gene. This gene functions as a transcription factor that represses expression of the γ-globin gene and hence inhibits the production of fetal hemoglobin in adult erythrocytes. Through precise targeting and the creation of a double-stranded break (DSB) in the BCL11A gene using CRISPR-Cas9 technology, this process facilitates the expression of γ-globin in edited cells through the inactivation of BCL11A7.
To achieve the desired edit in the genomes of a patient’s erythrocytes, autologous hematopoietic stem and progenitor cells (HSPCs) are extracted from the patient’s bone marrow. The HSPCs are genetically edited in vitro via the CRISPR-Cas9 system to reactivate the production of fetal hemoglobin. These cells are then infused back into the patient. The edited HPSCs give rise to fetal hemoglobin producing erythrocytes.
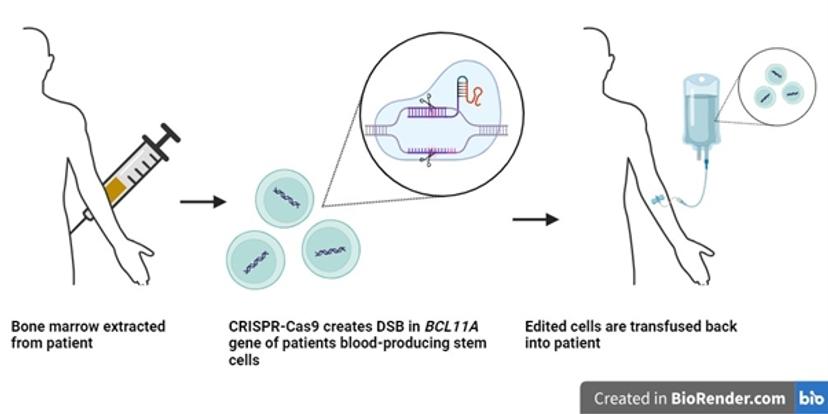
The therapy has been shown to yield clinically meaningful and sustained improvements in hemoglobin levels. This has eliminated the need for transfusion and prevented vaso-occlusive episodes in patients treated with Casgevy, effectively curing patients of the disease at the molecular level8.
The challenges
Casgevy harnessed CRISPR technology to modify patient’s cells in vitro for return to the body. However, this in vitro approach is not applicable to most disease types9.
Stability, efficiency, and safety concerns of the in vivo use of CRISPR-Cas9 technology have hindered its clinical applicability thus far. This includes the potential to induce off-target modifications, stimulate the immune response, and the current lack of an efficient delivery approach10. Solving these challenges of in vivo CRISPR therapeutics is essential to realize the full potential of CRISPR as a commonplace treatment modality.
Immunogenicity
In vivo delivery of CRISPR therapeutics bears immunogenicity challenges as every component has been found to activate the immune system. CRISPR effectors, such as Cas9, can prompt specific effector and memory adaptive immune responses as they are large foreign proteins derived from bacteria or archaea11. Cytotoxicity can result from the secondary structure of gRNAs through its activation of an innate immune response mediated by pattern recognition receptor proteins. Resolving this issue through modification of the gRNA poses a challenge, as the hairpin secondary structures are important to effectively engage Cas proteins and to maintain intracellular stability of the gRNA12. Sadly, a man died in one clinical trial as a result of a strong immune response to a high dose of adeno-associated virus vector. The trial was designed to assess the effectiveness of a CRISPR-Cas9 therapy intended for the treatment of Duchenne muscular dystrophy13.
Off-target modifications
Off-target effects remain a significant hurdle to the safe integration of CRISPR therapeutics into clinical applications. Off-target effects arise when Cas9 acts on non-targeted genomic sites, resulting in cleavages that may lead to unintended and adverse outcomes14. Current computational tools have traditionally shown limited performance in genome-wide off-target prediction, due to lack of comprehensive understanding of the CRISPR molecular mechanism. However, ongoing research is integrating molecular dynamics simulations to enhance the precision of computational analysis, establishing a framework for more accurate genome-wide CRISPR off-target prediction and sgRNA optimization15. Although promising, research indicates that off-target outcomes in therapeutic gene editing are influenced by variations in human genetic diversity. Given the reliance of standard computational tools and biochemical methods on reference genomes, this may suggest that a focus towards variant aware off-target assessment will become crucial to therapeutic genome editing evaluation and offer a powerful approach for comprehensive off-target nomination16.
Stability
In vivo gene therapy products require the use of delivery vectors to prevent immediate degradation and immune clearance. Adeno-associated virus (AAV) vectors are the most commonly used for delivering gene drugs, however, pose limitations for widespread application in disease due to restricted loading capacity, a deficiency in specific targeting ability, and an inability to integrate into the host genome.
Non-viral vectors have been gaining popularity in research in recent years for use in CRISPR therapeutics, including lipid nanoparticles, polymeric nanoparticles, biomimetic nanomaterials, and exosomes. Non-viral vectors with high loading capacities and good safety profiles, like lipid nanoparticles (LNPs), have emerged as excellent choices for delivery, and have been used in clinical trials. However, LNPs frequently elicit immunogenic responses in vivo. On the other hand, exosomes derived from organisms or biofilms prove highly effective in evading immune clearance when utilized as non-viral vector masks. Additionally, the proteins and peptides enriched in the biofilm can facilitate the targeted delivery of gene drugs to specific cells17.
Safety
There have been concerns of the DNA-damage toxicity of CRISPR therapeutics as CRISPR/Cas9 mechanism causes DSBs. This is due to reports of p53 induced apoptosis in response to DSBs introduced by CRISPR. Additionally, on-target large deletions spanning kilobases and complex rearrangements can be unintended consequences, highlighting a significant safety concern for clinical applications18.
Variations of Cas9 which do not induce DSBs have been developed and may provide promise for therapeutic application. For example, dCas9 facilitates transient gene expression manipulation by fusing transcriptional activating or repressing domains or proteins to the DNA-binding effector, avoiding the introduction of DSBs. Cas9 nickase (Cas9n) is another altered Cas9 protein and induces single-stranded breaks rather than DSBs19. These altered Cas9 proteins has given rise to novel and powerful approaches; base editing and prime editing. These approaches accurately incorporate point mutations without the need for DSBs or a DNA donor template20.
References:
SelectScience. (2023, November 16). Dr. Priya Chockalingam. [online] The Scientists’ Channel®, available at: https://thescientistschannel.com/priya-chockalingam.
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J.A. and Charpentier, E. (2012). A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science, 337(6096), 816–821. https://doi.org/10.1126/science.1225829.
Medicines and Healthcare products Regulatory Agency. (2023, November 16). MHRA authorises world-first gene therapy that aims to cure sickle-cell disease and transfusion-dependent β-thalassemia. GOV.UK, available at: https://www.gov.uk/government/news/mhra-authorises-world-first-gene-therapy-that-aims-to-cure-sickle-cell-disease-and-transfusion-dependent-thalassemia.
Doudna, J.A. (2020). The promise and challenge of therapeutic genome editing. Nature, 578(7794), 229–236. https://doi.org/10.1038/s41586-020-1978-5.
Asmamaw, M. and Zawdie, B. (2021). Mechanism and Applications of CRISPR/Cas-9-Mediated Genome Editing. Biologics: Targets & Therapy, 15, 353–361. https://doi.org/10.2147/BTT.S326422.
Li, T., Yang, Y., Qi, H., Cui, W., Zhang, L., Fu, X., He, X., Liu, M., Li, P. and Yu, T. (2023). CRISPR/Cas9 therapeutics: progress and prospects. Signal Transduction and Targeted Therapy, 8(36), 1–23. https://doi.org/10.1038/s41392-023-01309-7.
Canver, M. C., Smith, E. C., Sher, F., Pinello, L., Sanjana, N. E., Shalem, O., Chen, D. D., Schupp, P. G., Vinjamur, D. S., Garcia, S. P., Luc, S., Kurita, R., Nakamura, Y., Fujiwara, Y., Maeda, T., Yuan, G.-C., Zhang, F., Orkin, S. H., & Bauer, D. E. (2015). BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature, 527(7577), 192–197. https://doi.org/10.1038/nature15521.
Frangoul, H., Altshuler, D., Cappellini, M. D., Chen, Y.-S., Domm, J., Eustace, B. K., Foell, J., de la Fuente, J., Grupp, S., Handgretinger, R., Ho, T. W., Kattamis, A., Kernytsky, A., Lekstrom-Himes, J., Li, A. M., Locatelli, F., Mapara, M. Y., de Montalembert, M., Rondelli, D., & Sharma, A. (2021). CRISPR-Cas9 Gene Editing for Sickle Cell Disease and β-Thalassemia. New England Journal of Medicine, 384(3), 252–260. https://doi.org/10.1056/nejmoa2031054.
Li, T., Yang, Y., Qi, H., Cui, W., Zhang, L., Fu, X., He, X., Liu, M., Li, P., & Yu, T. (2023). CRISPR/Cas9 therapeutics: progress and prospects. Signal Transduction and Targeted Therapy, 8(1), 1–23. https://doi.org/10.1038/s41392-023-01309-7.
Rasul, M. F., Hussen, B. M., Salihi, A., Ismael, B. S., Jalal, P. J., Zanichelli, A., Jamali, E., Baniahmad, A., Ghafouri-Fard, S., Basiri, A., & Taheri, M. (2022). Strategies to overcome the main challenges of the use of CRISPR/Cas9 as a replacement for cancer therapy. Molecular Cancer, 21(1). https://doi.org/10.1186/s12943-021-01487-4.
Ewaisha, R. and Anderson, K.S. (2023). Immunogenicity of CRISPR therapeutics—Critical considerations for clinical translation. Frontiers in Bioengineering and Biotechnology, 11. https://doi.org/10.3389/fbioe.2023.1138596.
Chew, W. L. (2017). Immunity to CRISPR Cas9 and Cas12a therapeutics. Wiley Interdisciplinary Reviews: Systems Biology and Medicine, 10(1), e1408. https://doi.org/10.1002/wsbm.1408.
Philippidis, A. (2023). Case Study Cites Immune Reaction to High Adeno-Associated Virus Dose in Explaining Duchenne Muscular Dystrophy Trial Death. Human Gene Therapy, 34(13-14), 588–591. https://doi.org/10.1089/hum.2023.29244.bfs.
Guo, C., Ma, X., Gao, F. and Guo, Y. (2023). Off-target effects in CRISPR/Cas9 gene editing. Frontiers in Bioengineering and Biotechnology, 11:1143157. https://doi.org/10.3389/fbioe.2023.1143157.
Chen, Q., Chuai, G., Zhang, H., Tang, J., Duan, L., Guan, H., Li, W., Li, W., Wen, J., Zuo, E., Zhang, Q., & Liu, Q. (2023). Genome-wide CRISPR off-target prediction and optimization using RNA-DNA interaction fingerprints. Nature Communications, 14(1), 7521. https://doi.org/10.1038/s41467-023-42695-4.
Cancellieri, S., Zeng, J., Lin, L. Y., Tognon, M., Nguyen, M. A., Lin, J., Bombieri, N., Maitland, S. A., Ciuculescu, M.-F., Katta, V., Tsai, S. Q., Armant, M., Wolfe, S. A., Giugno, R., Bauer, D. E., & Pinello, L. (2023). Human genetic diversity alters off-target outcomes of therapeutic gene editing. Nature Genetics, 55(1), 34–43. https://doi.org/10.1038/s41588-022-01257-y.
Li, T., Yang, Y., Qi, H., Cui, W., Zhang, L., Fu, X., He, X., Liu, M., Li, P. and Yu, T. (2023). CRISPR/Cas9 therapeutics: progress and prospects. Signal Transduction and Targeted Therapy, 8(1), 1–23. https://doi.org/10.1038/s41392-023-01309-7.
Uddin, F., Rudin, C. M., & Sen, T. (2020). CRISPR Gene Therapy: Applications, Limitations, and Implications for the Future. Frontiers in Oncology, 10(1387). https://doi.org/10.3389/fonc.2020.01387.
Anzalone, A. V., Randolph, P. B., Davis, J. R., Sousa, A. A., Koblan, L. W., Levy, J. M., Chen, P. J., Wilson, C., Newby, G. A., Raguram, A., & Liu, D. R. (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature, 576(7785), 149–157. https://doi.org/10.1038/s41586-019-1711-4.
Tao, J., Bauer, D.E. & Chiarle, R. (2023). Assessing and advancing the safety of CRISPR-Cas tools: from DNA to RNA editing. Nat Communications, 14, 212. https://doi.org/10.1038/s41467-023-35886-6.
