Antimicrobial resistance and the MRSA crisis: A silent pandemic demanding urgent global action
In this guest editorial, Microzone explores MRSA and the escalating crisis of antimicrobial resistance—demanding faster diagnostics, renewed stewardship, and urgent global action
18 Jun 2025
Antimicrobial resistance (AMR) is no longer a looming threat—it's a present-day crisis. With over 1.27 million deaths directly attributable to AMR in 2019, and nearly 5 million associated fatalities globally according to the GRAM study published in The Lancet (Murray et al., 2022), the statistics are sobering. In the United Kingdom alone, an estimated 7,600 deaths were directly linked to AMR that year. These numbers are not abstract—they are families affected, healthcare systems strained, and economic productivity diminished.
Among the most notorious contributors to this resistance is methicillin-resistant Staphylococcus aureus (MRSA)—a pathogen that has carved out an enduring place in hospital wards and community settings alike. What makes MRSA particularly insidious is its adaptability: once a hospital-associated problem, it now flourishes in the community, increasingly affecting younger and healthier individuals.
Why MRSA still matters
Discovered in the early 1960s, MRSA emerged soon after methicillin itself. It differs from methicillin-susceptible S. aureus (MSSA) due to the acquisition of the mecA or mecC genes, which encode for an alternative penicillin-binding protein (PBP2a) that renders β-lactam antibiotics ineffective (figure 1). This genetic sleight of hand makes treatment options more limited, more expensive, and often more deadly.
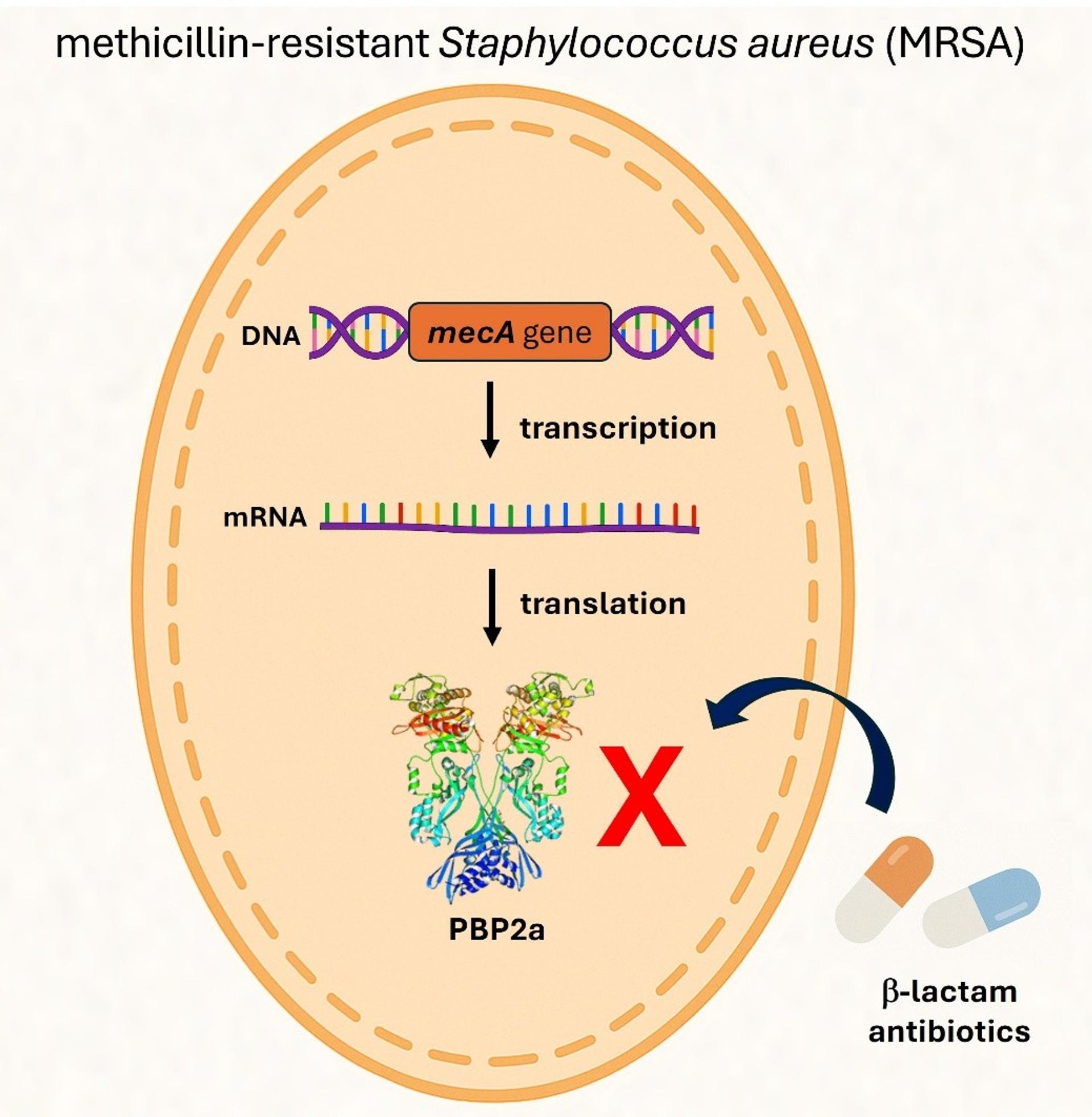
Figure 1. Genetic Mechanism of the mecA Gene in MRSA. The mecA gene encodes a modified penicillin-binding protein known as PBP2a. Unlike the native PBPs targeted by β-lactam antibiotics, PBP2a has a significantly lower affinity for these drugs. β-lactam antibiotics—including methicillin—normally inhibit bacterial growth by binding to PBPs and disrupting cell wall synthesis. However, the presence of PBP2a allows Staphylococcus aureus to bypass this inhibition, enabling continued cell wall synthesis and survival even in the presence of β-lactam antibiotics.
In 2023, Shoaib et al. reiterated the global clinical burden of MRSA, with particular emphasis on severe outcomes like bloodstream infections and pneumonia. UK-specific studies like those by Said et al. (2025) and Lawes et al. (2015) underscore MRSA's entrenched role in ICU morbidity and hospital-acquired infections—despite decades of control strategies including admission screening and decolonisation protocols.
Diagnostics: The double-edged sword
While much progress has been made in MRSA diagnostics, the tools themselves present both solutions and limitations. Culture-based methods remain the gold standard for phenotypic susceptibility testing, offering valuable antimicrobial guidance. Yet, they are slow—results can take up to 72 hours, which delays critical infection control decisions.
Molecular diagnostics, particularly real-time PCR (qPCR), offer a compelling alternative. Kits like the Microzone MRSA qPCR Detection Kit exemplify how modern platforms can rapidly identify MRSA by detecting both methicillin resistance genes (mecA and mecC) and species-specific markers (femA and nuc) in under an hour. The Microzone kit also includes an internal amplification control (IAC) to ensure test integrity and employs UNG chemistry to minimise false positives due to contamination. Its flexibility in accepting either extracted DNA or direct-to-PCR lysates makes it ideal for both routine clinical and high-throughput surveillance applications.

Choosing the right diagnostic method depends largely on the clinical setting and intended purpose. Molecular-based diagnostics provide a versatile solution across diverse clinical scenarios. In high-volume laboratories, prioritise assays that support scalability, automation, and seamless integration with laboratory information systems. For point-of-care or rapid triage, focus on speed and simplicity, favouring closed-cartridge systems that require minimal hands-on time. In cases of outbreak control and epidemiological tracking, select platforms with validated surveillance capabilities and robust resistance gene detection. However, when it comes to guiding treatment decisions, culture-based methods remain indispensable due to their ability to provide phenotypic susceptibility data.
The cost of inaction
We are long past the era where AMR, and by extension MRSA, can be dismissed as a niche or technical issue. AMR is a global health emergency on par with pandemics and climate change. The UK government’s own projections suggest 10 million annual deaths globally by 2050 if current trajectories remain unchanged and the cumulative economic impact could reach $100 trillion USD, severely undermining global productivity. To put the scale in perspective, by 2050, AMR could be responsible for one death every three seconds (UK Review on Antimicrobial Resistance, 2016).
The way forward demands coordinated global surveillance, investment in rapid diagnostics, stewardship of existing antibiotics, and an unrelenting push for new drug development. It also requires public engagement—AMR is not merely a hospital problem. It’s an agricultural problem, an environmental problem, and above all, a societal problem (figure 2).
Unless we act decisively, MRSA and other drug-resistant pathogens will not remain clinical curiosities—they will define the future of global health.
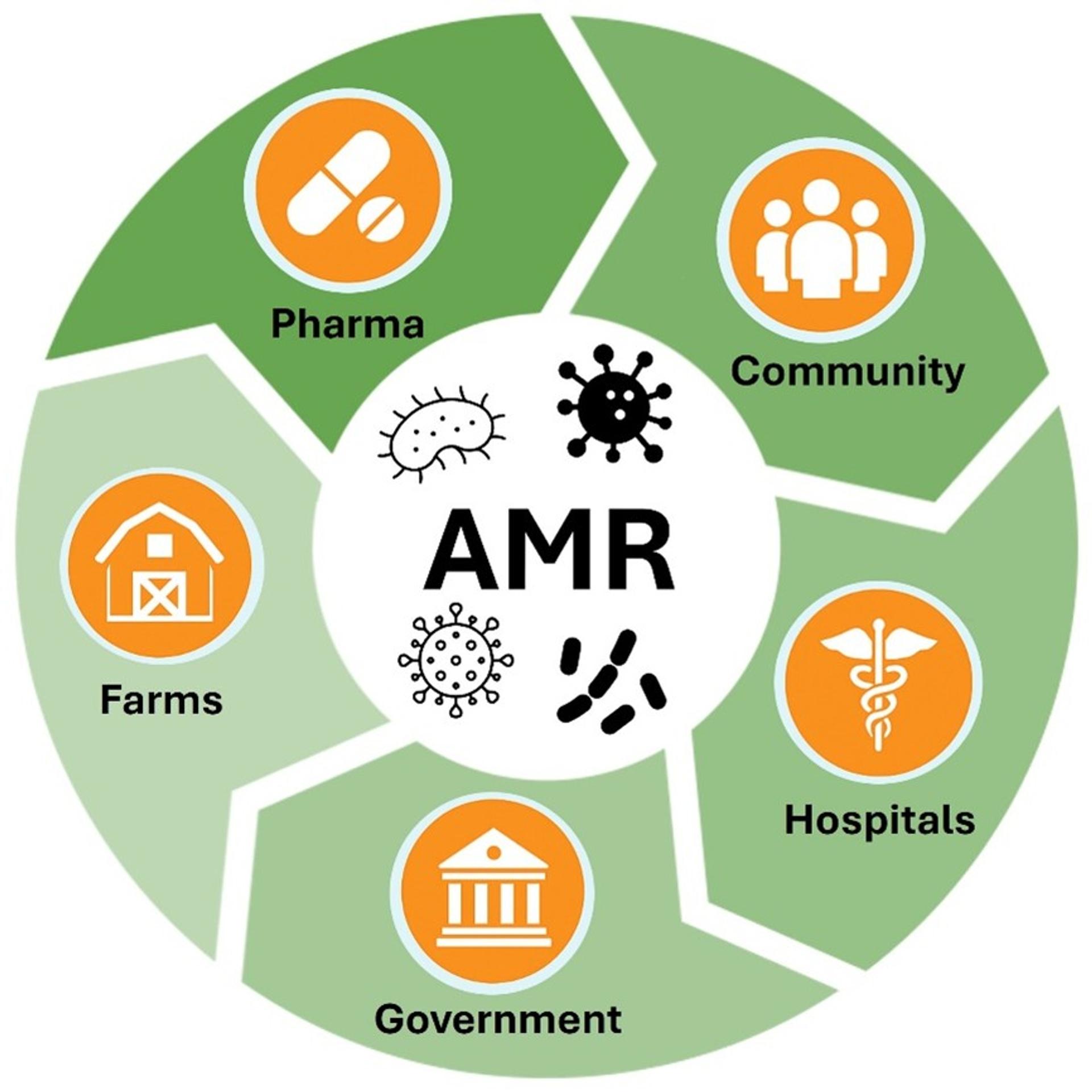
Figure 2. Antimicrobial resistance (AMR) is a shared societal challenge.
AMR requires broad public engagement as it is not solely a hospital-based issue, but also deeply rooted in agriculture, environmental contamination, pharmaceutical practices, community and public health policy.
References:
O’Neill, J. (2016) Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. Review on Antimicrobial Resistance. Wellcome Trust and HM Government.
Lawes, T et al. (2015) Effects of national antibiotic stewardship and infection control strategies on hospital-associated and community-associated methicillin-resistant Staphylococcus aureus infections across a region of Scotland: a non-linear time-series study. Lancet Infect Dis. Dec;15(12):1438-49.
Murray, C J L et al. (2022) Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. The Lancet, Volume 399, Issue 10325, 629 – 655.
Said, K B et al. (2025) MRSA Profiles Reveal Age- and Gender-Specificity in a Tertiary Care Hospital: High Burden in ICU Elderly and Emerging Community Patterns in Youth. Microorganisms. May 6;13(5):1078.
Shoaib, M et al. (2023) MRSA compendium of epidemiology, transmission, pathophysiology, treatment, and prevention within one health framework. Front Microbiol. Jan 10;13:1067284.