Resources
25
Application Notes
Validated stains for spheroids, organoids, and other 3D cultures
Product Brochures
witec360 confocal Raman microscope
Product Brochures
The special one: AIA-360 immunoassay analyzer
Application Notes
Freeze-drying illustrated laboratory guide
Application eBooks
Miniaturize to maximize: Precision liquid handling & dispensing for discovery and applied research
Essential techniques for PCR, mass spec sample prep, direct titration, assay development, and more
Application eBooks
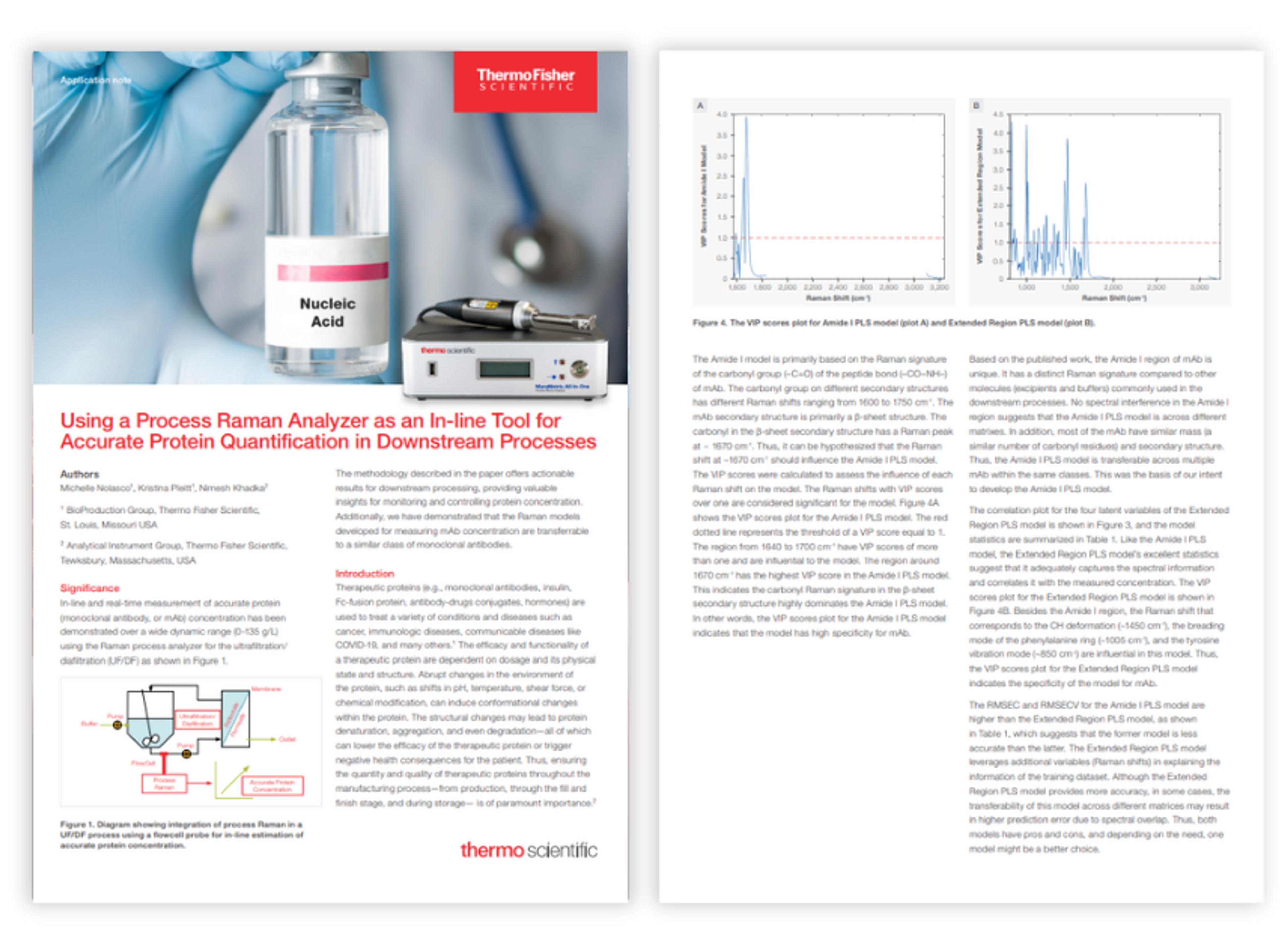
Transforming bioprocess data into actionable intelligence
Product Brochures
The compact HbA1c analyzer with big impact
Application eBooks
Streamline liquid handling with automation
Product Brochures
Introducing the Omnia Discovery
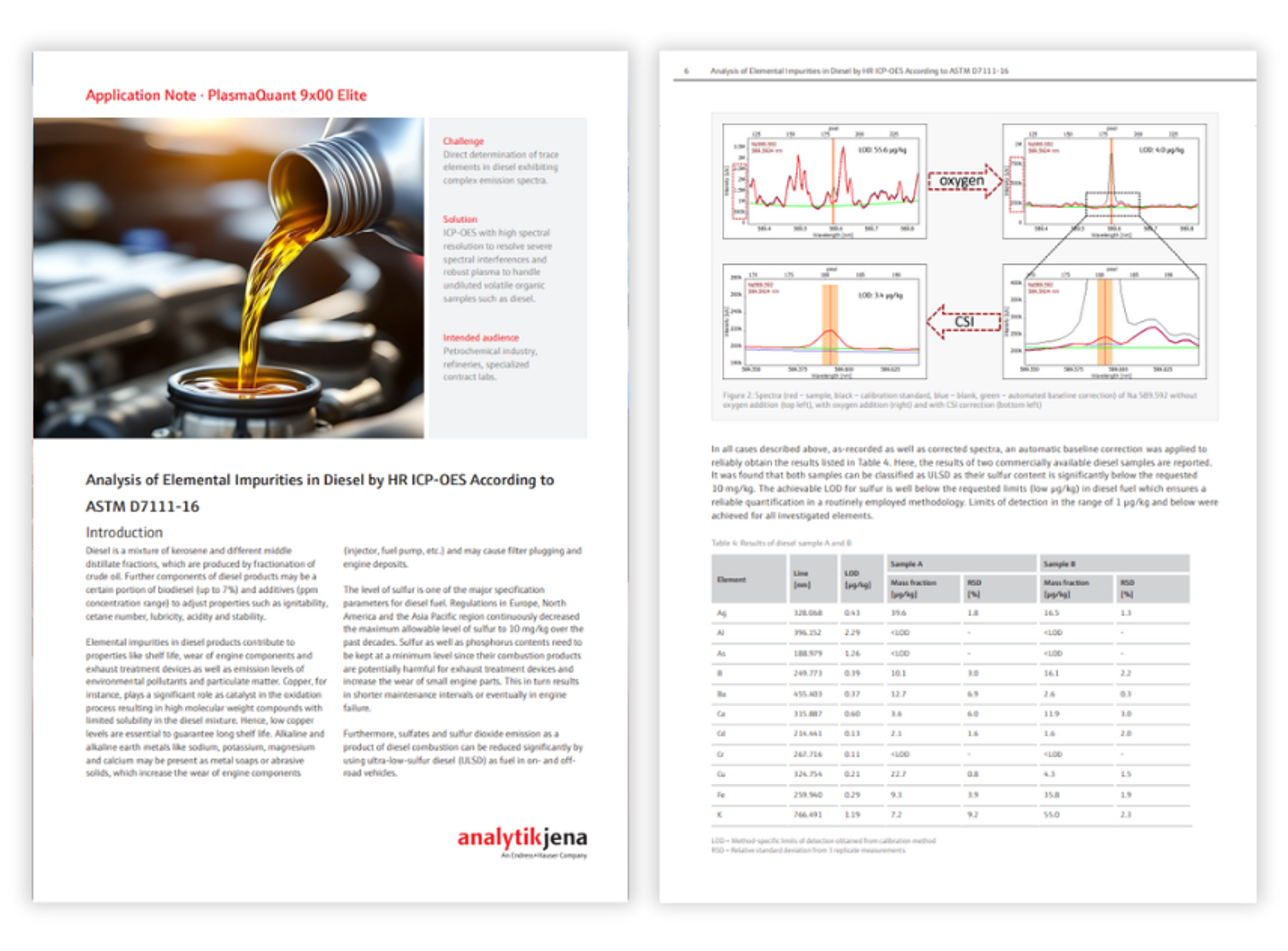
Application Notes
Food microbiology testing methods for Listeria species
Application Notes
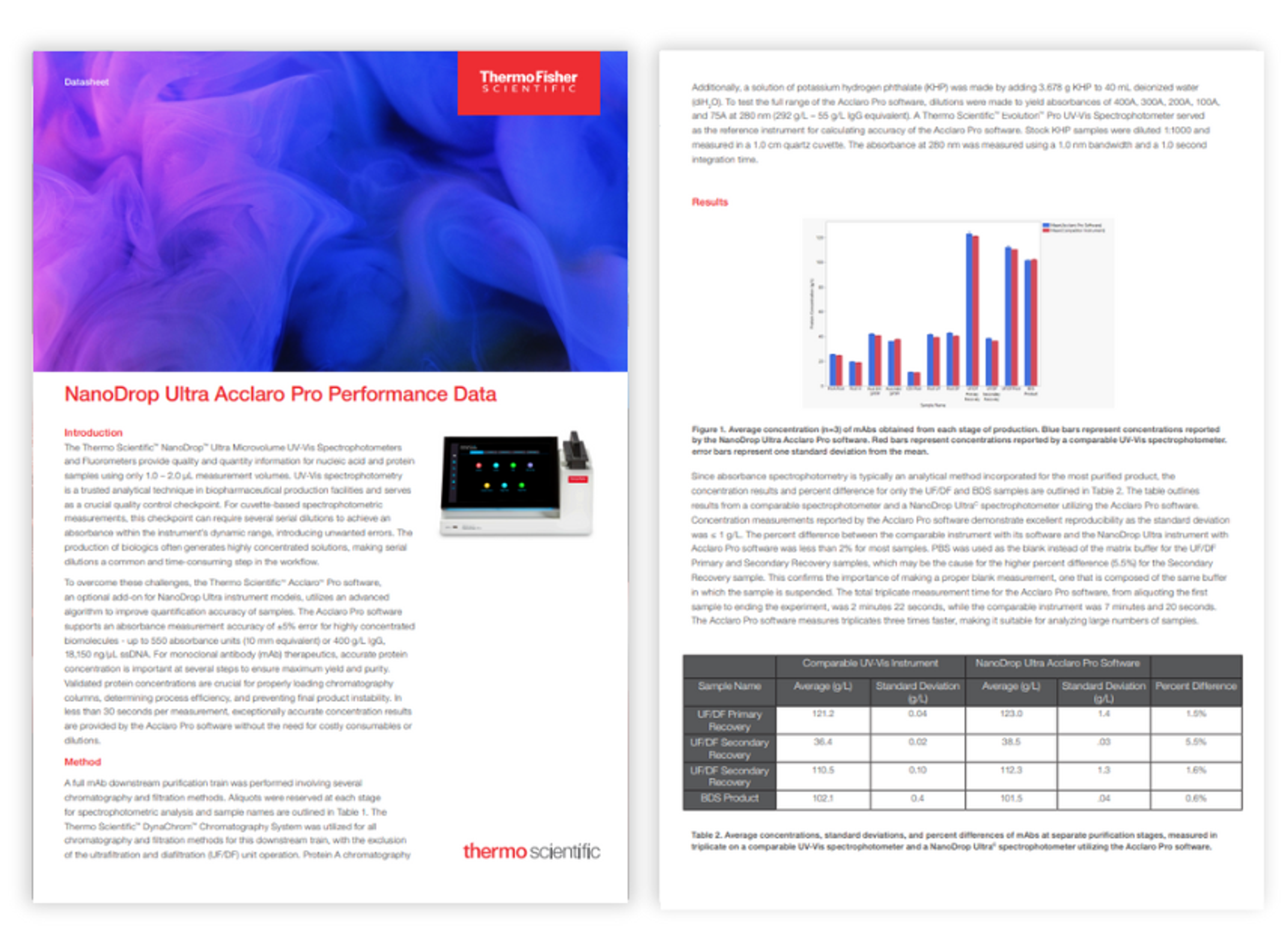
NanoDrop Ultra Acclaro Pro performance data
Application Notes
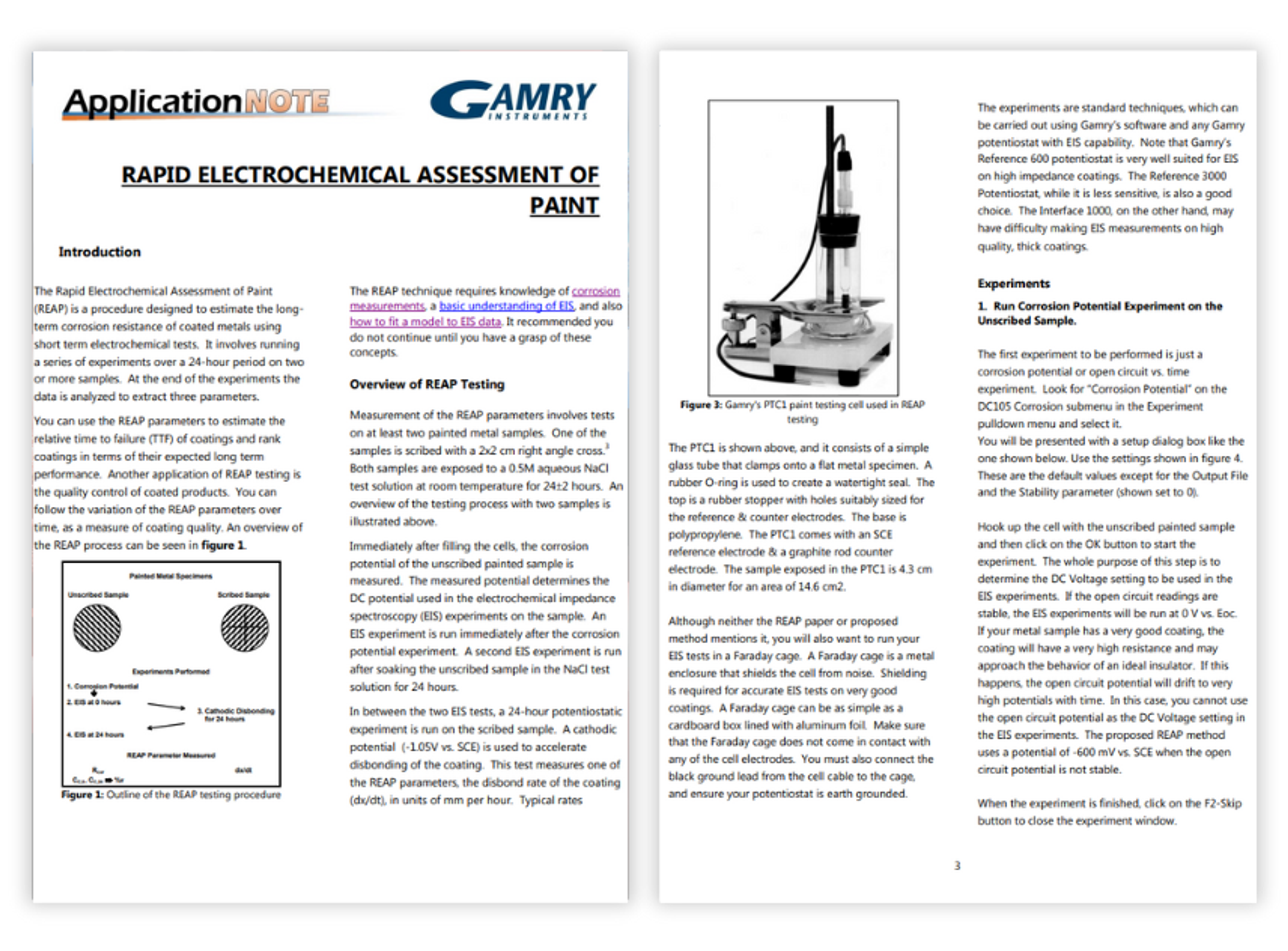
Rapid electrochemical assessment of paint
Product Brochures